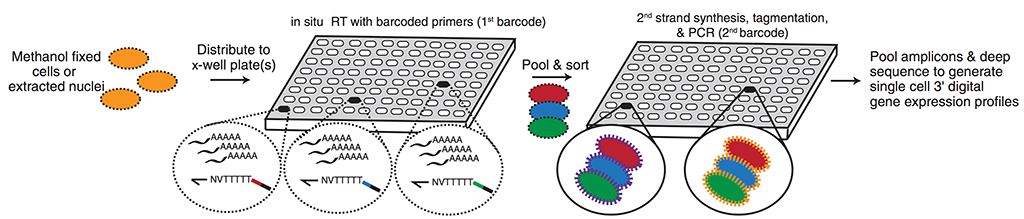
Single-cell RNA-Seq reports the mRNA abundances of every gene in the genome in many individual cells in a single experiment. In collaboration with Jay Shendure’s lab and scientists at Illumina, we recently developed sci-RNA-seq, which uses combinatorial cellular indexing, captures transcriptomes for tens of thousands of cells in a single experiment for a fraction of the cost of alternative methods. The technique scales sublinearly in time and cost, enabling the profiling of all the cells in whole animals.