Single-cell genomics offers a means of precisely quantifying the state of individual cells and thus may enable the construction of explicit, genome-scale dynamical cellular models. Early single-cell transcriptomic studies lend support to the idea that cells are occupants of a vast, complex landscape of possible states and raise doubts that cell types are precisely defined, discrete entities. Time series experiments of differentiation have observed cells transitioning between a starting state and one or more end states, with many cells distributed along a “trajectory” between them. The Monocle algorithm introduced the notion of pseudotime, a quantitative measure of biological progression through a process such as cell differentiation.
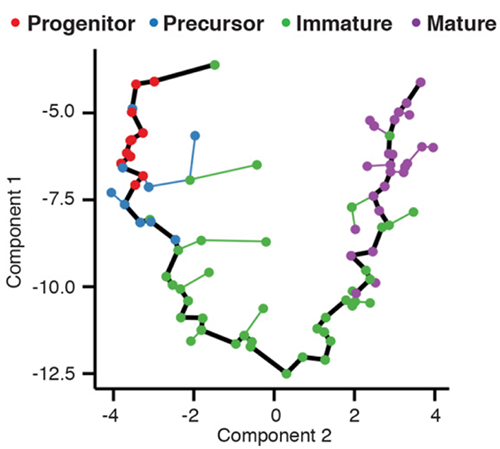
The trajectory plot above shows the trajectory followed by olfactory neurons as the develop in mice. Each point is a cell, where are connected into a minimum spanning tree, the core data structure Monocle uses to find the trajectory, shown in black. Each cell’s pseudotime value is measured as the distance along the trajectory from its position back to the beginning. In order to describe complex differentiation processes in which cells make fate decisions, Monocle allows these trajectories to have a branched structure with multiple possible outcomes.
The Trapnell laboratory is currently focused on three ways to extend single-cell trajectory analysis:
- Methods to identify genes that are differentially regulated along the trajectory, particularly near branches
- Algorithms for distinguishing “driver” genes that regulate progress along a trajectory or govern branching
- Building trajectories from new single-cell data types, such as single-cell ATAC-Seq.