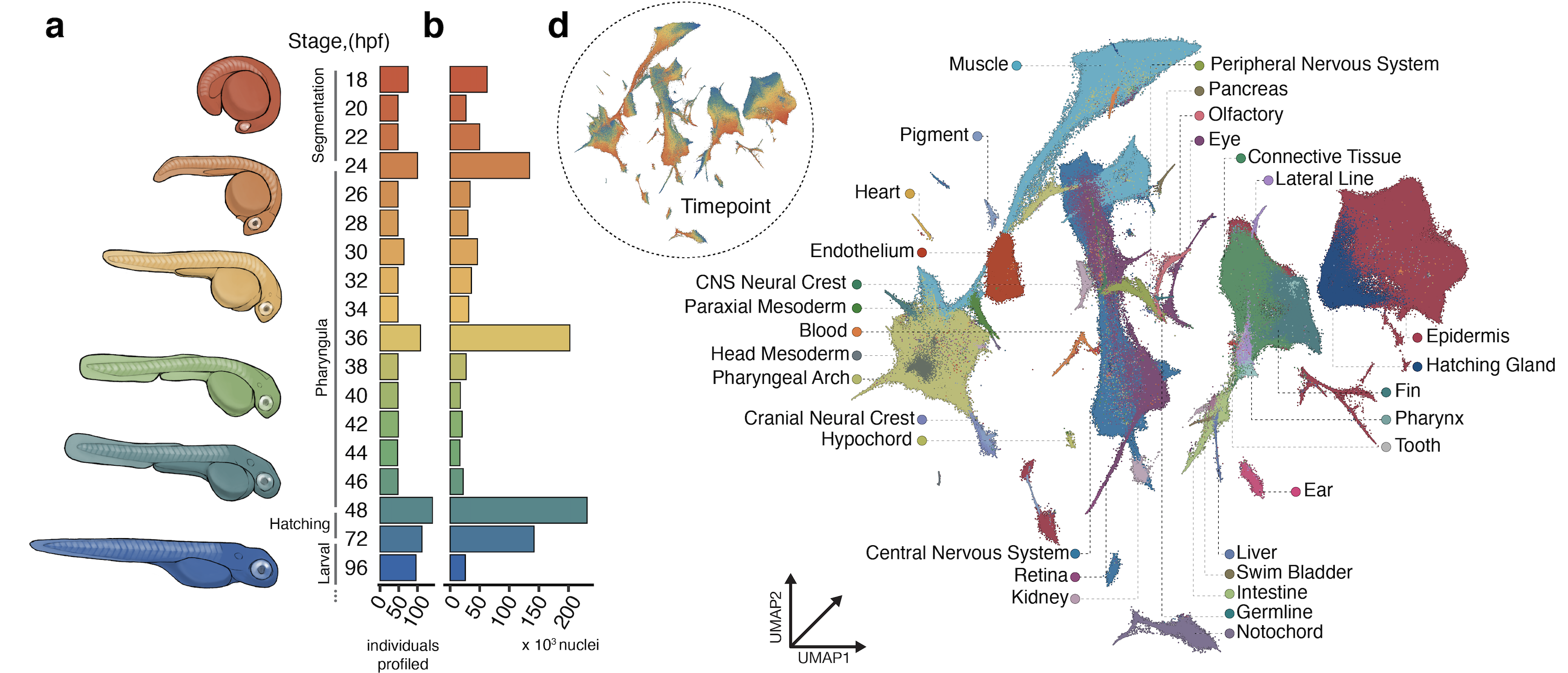
Exponential advances in throughput of single cell genomics technologies have led to a watershed moment in developmental biology. We can now feasibly and economically sequence millions of cells from thousands of specimens, which allows us to interrogate the effects of diverse genetic, chemical, and environmental perturbations on embryonic development. In effect, we can study gene regulation across individuals at whole-embryo scale and single-cell resolution to see how a perturbation impacts every gene, in every cell type, at every stage of its development.
We are working to systematically perturb zebrafish embryonic development and read out the effects using massively multiplexed single-cell molecular profiling. These experiments will not only facilitate the mapping of the gene networks that control vertebrate development, but also will shed light on how those networks buffer variability and confer robustness. Such a map would dramatically accelerate efforts to understand the molecular mechanisms that determine severity in a wide range of human developmental disorders.
