Abstract
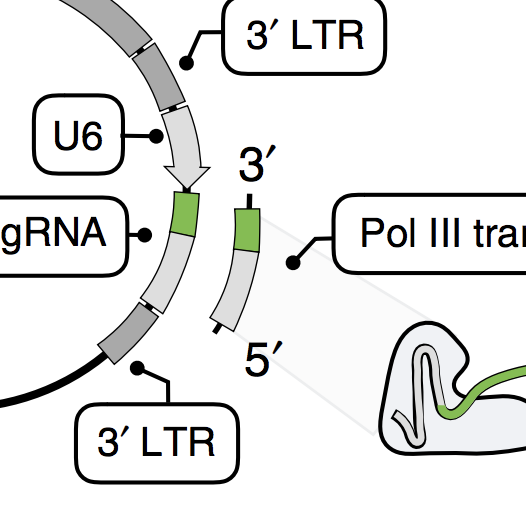
Several groups recently coupled CRISPR perturbations and single-cell RNA-seq for pooled genetic screens. We demonstrate that vector designs of these studies are susceptible to ~50% swapping of guide RNA–barcode associations because of lentiviral template switching. We optimized a published alternative, CROP-seq, in which the guide RNA also serves as the barcode, and here confirm that this strategy performs robustly and doubled the rate at which guides are assigned to cells to 94%.
*co-first author
**co-senior author