Abstract
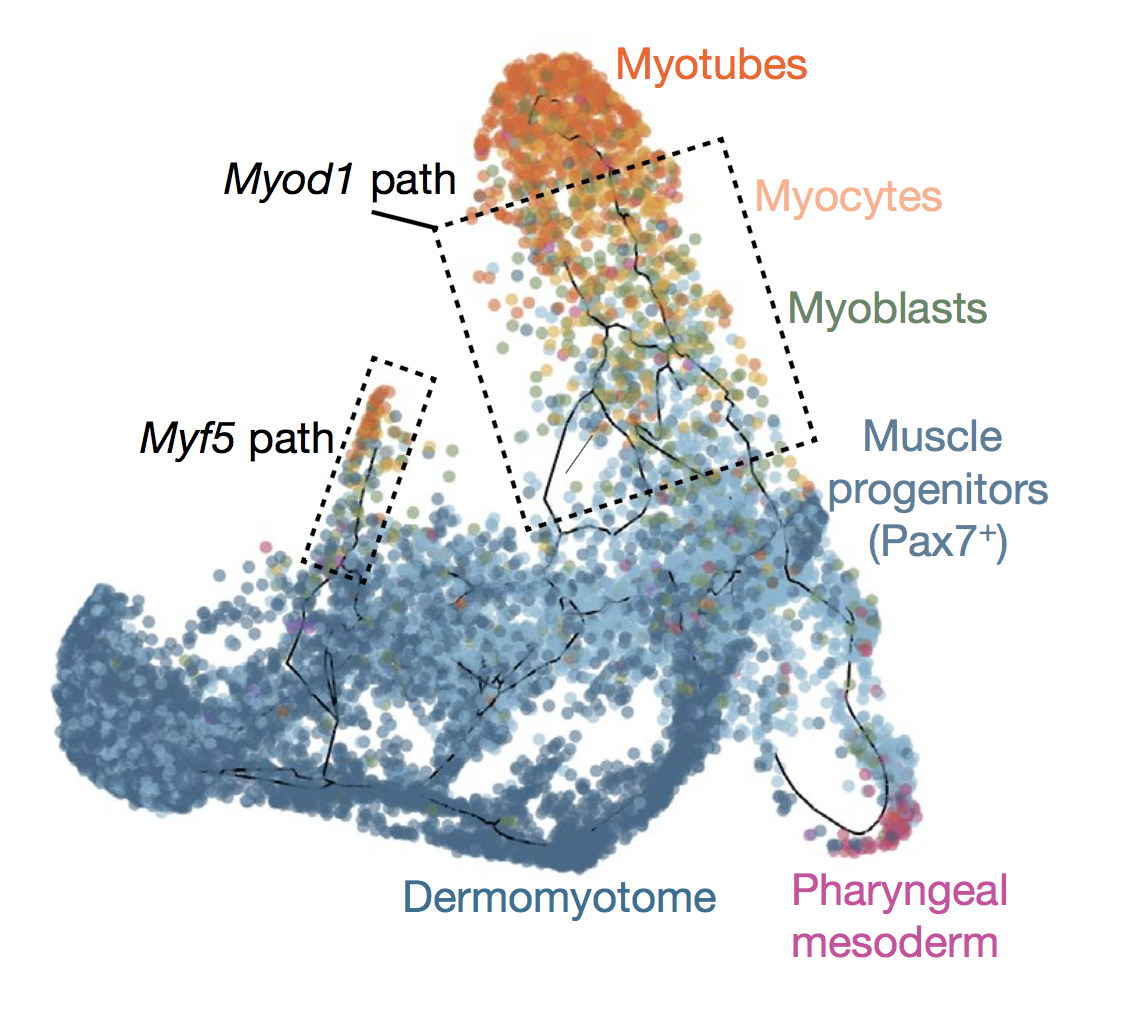
Mammalian organogenesis is a remarkable process. Within a short timeframe, the cells of the three germ layers transform into an embryo that includes most of the major internal and external organs. Here we investigate the transcriptional dynamics of mouse organogenesis at single-cell resolution. Using single-cell combinatorial indexing, we profiled the transcriptomes of around 2 million cells derived from 61 embryos staged between 9.5 and 13.5 days of gestation, in a single experiment. The resulting ‘mouse organogenesis cell atlas’ (MOCA) provides a global view of developmental processes during this critical window. We use Monocle 3 to identify hundreds of cell types and 56 trajectories, many of which are detected only because of the depth of cellular coverage, and collectively define thousands of corresponding marker genes. We explore the dynamics of gene expression within cell types and trajectories over time, including focused analyses of the apical ectodermal ridge, limb mesenchyme and skeletal muscle.
* co-first authors
** co-corresponding authors