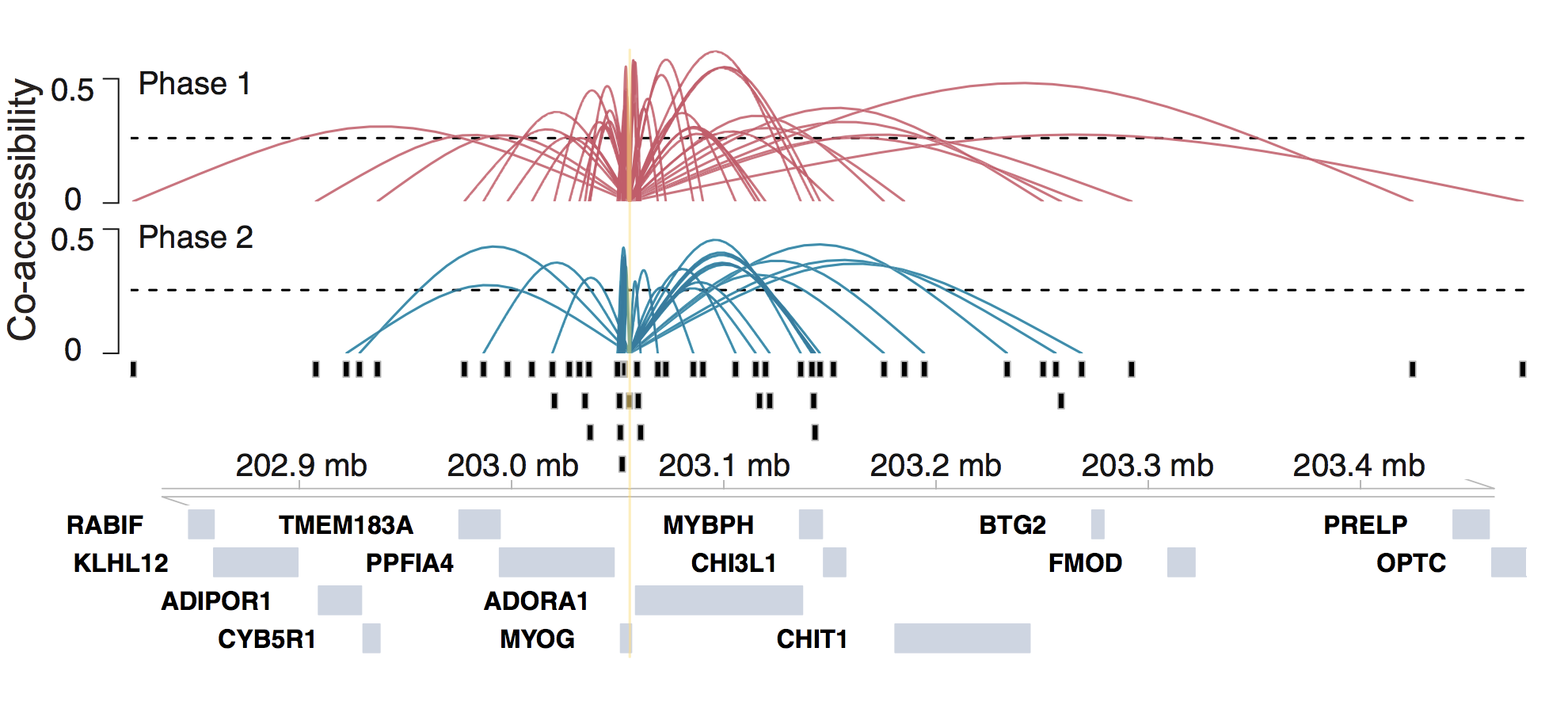
Cicero is an R package that provides tools for analyzing single-cell chromatin accessibility experiments. The main function of Cicero is to use single-cell chromatin accessibility data to predict cis-regulatory interactions (such as those between enhancers and promoters) in the genome by examining co-accessibility. In addition, Cicero extends Monocle to allow clustering, ordering, and differential accessibility analysis of single cells using chromatin accessibility. For more information about Cicero, explore our publications.
Cicero is a collaborative effort between Cole Trapnell’s lab and Jay Shendure’s lab. It was written by Hannah Pliner.