Trajectories
Build single-cell trajectories with the software that introduced pseudotime. Find cell fate decisions and the genes regulated as they're made.

Clustering
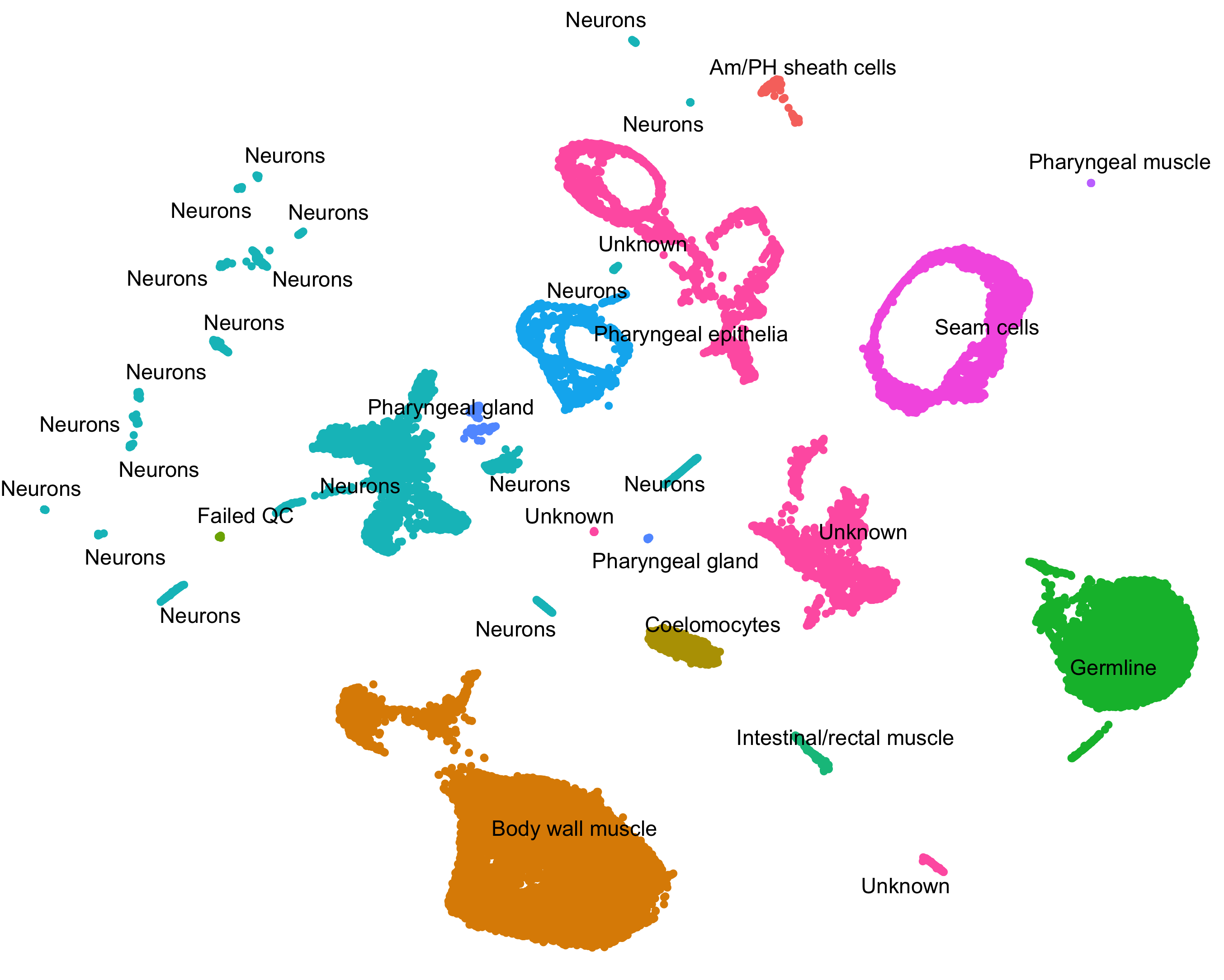
Group and classify your cells based on gene expression. Identify new cell types and states and the genes that distinguish them.

Differential expression
Find genes that vary between cell types and states, over trajectories, or in response to perturbations using statistically robust, flexible differential analysis.
Classify and count cells.
Single-cell RNA-Seq experiments allow you to discover (and possibly rare) subtypes of cells. Monocle 3 helps you identify them.
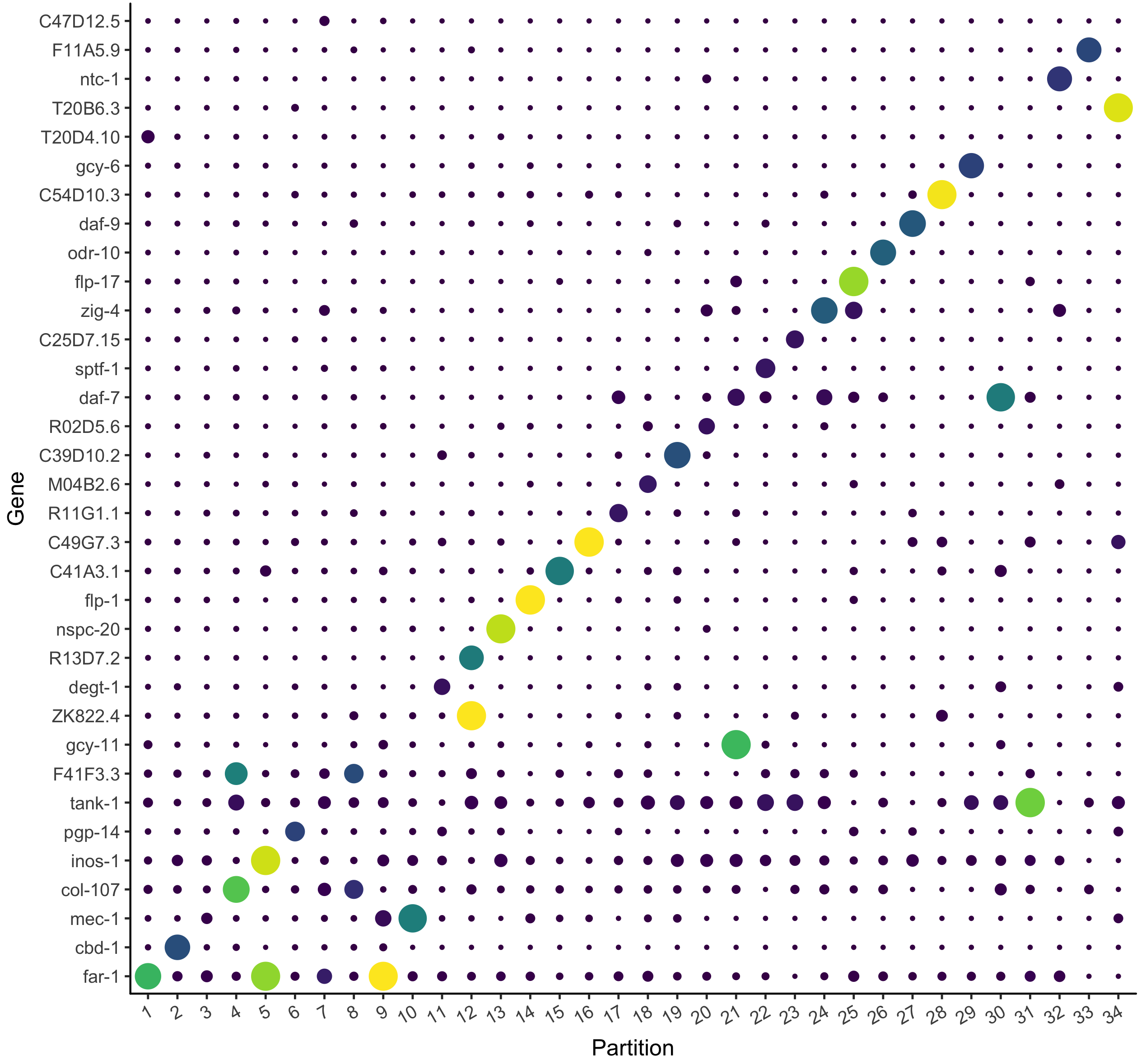
Identify new marker genes.
Many researchers are using single-cell RNA-Seq to discover new cell types. Monocle 3 can help you purify them or characterize them further by identifying key marker genes that you can use in follow up experiments such as immunofluorescence or flow sorting.
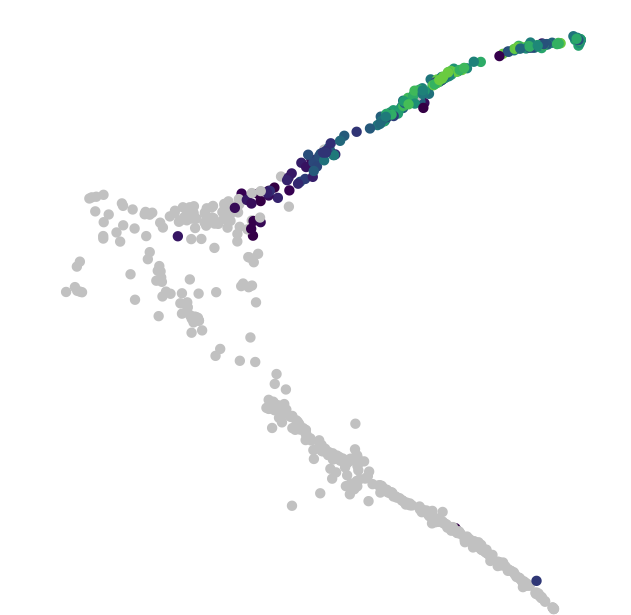
Robustly track changes over (pseudo) time.
In development, disease, and throughout life, cells transition from one state to another. Monocle introduced the concept of pseudotime, which is a measure of how far a cell has moved through biological progress.
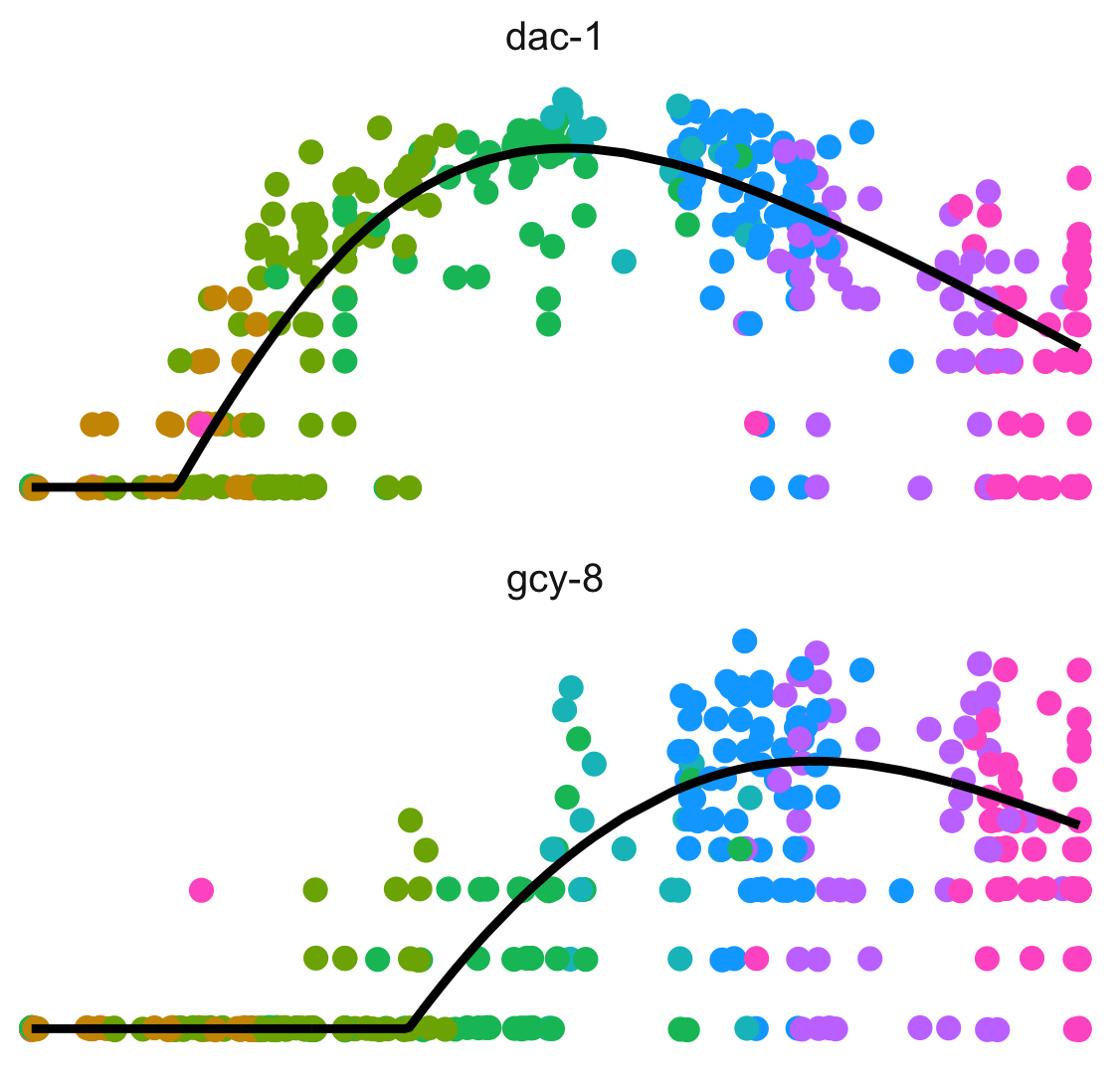
Dissect cellular decisions with branch analysis.
Single-cell trajectory analysis how cells choose between one of several possible end states. The new reconstruction algorithms introduced in Monocle 3 can robustly reveal branching trajectories, along with the genes that cells use to navigate these decisions.